Chapter 17 Exercise: Survival estimation
17.1 Introduction to mark-recapture modelling
Recapture or resightings (reencounters) of marked individuals inform about apparent survival, i.e. the probability that an individual survives and stays in the study area. Apparent survival differs from true survival because when the animals is seen last, it is still alive, i.e. the death is not observed. Thus, if an animal is no longer recaptured or resighted, we do not know whether it moved out of the study area or whether it died. Mark-recapture modelling techniques are designed to separately estimate recapture/resighting probability and apparent survival. To do so, they separate the observation process (recapturing or resighting an animal) from the biological process (staying in the study area and surviving). The basis for the development of an immense diversity of mark-recapture models has been set by the Cormack-Jolly-Seber model (Cormack 1964; Jolly 1965; Seber 1965). The CJS-model is introduced here using data on Snowfinches, a high alpine bird species that we study since 2015.
Since 2015, we individually marked 1310 Snowfinches. We systematically observed Snowfinches to read rings using scopes and cameras. We could resight 529 of the marked individuals in later years, many of them in more than one year. We collated the marking and resighting data in a capture history matrix \(y\). Each row of \(y\) contains the capture history of one individual and each column corresponds to a year from 2015 to 2023. A \(1\) indicates that the individual was resighted or recaptured during that year and a \(0\) indicates that the individual has not been seen. Thus, a \(1\) means the individual is alive in the study area and it has been detected by the researchers. A \(0\) could mean that the individual died, that it emigrated from the study area or that it is still alive in the study area but has not been detected by the researchers.
index <- order(datax$first)
ch <- datax$y[index,]
plot(c(1,datax$nocc), c(1, datax$nind), type="n", axes=FALSE, xlab="Yeaer", ylab="Individual")
axis(1, at=1:datax$nocc)
axis(2, at=1:datax$nind, las=1, cex.lab=0.5)
for(i in 1:datax$nind){
points(c(1:datax$nocc)[ch[i,]==1], rep(i,sum(ch[i,])), cex=.6)
lines(c(1:datax$nocc)[ch[i,]==1], rep(i,sum(ch[i,])))
}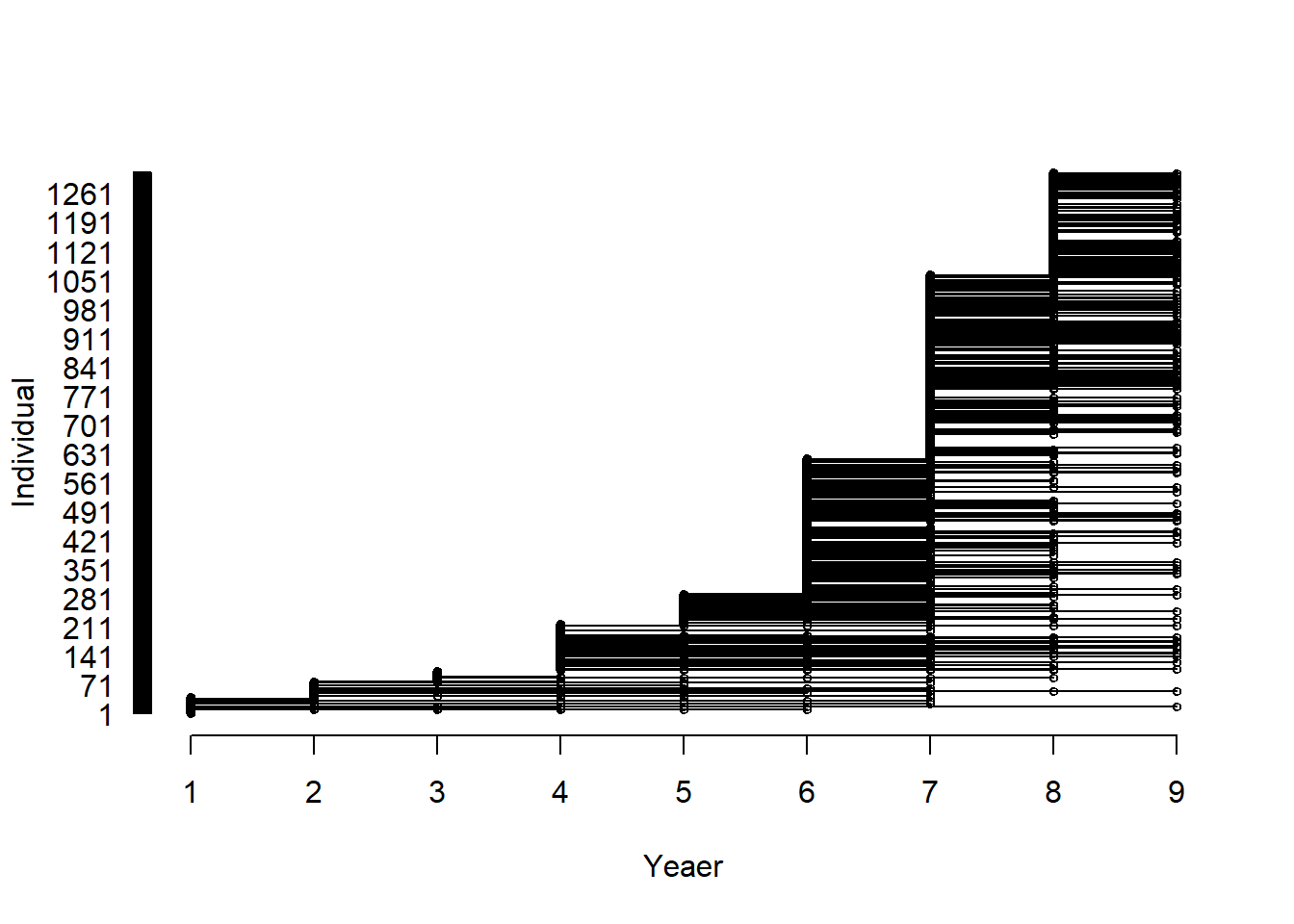
Figure 17.1: Visualisation of the capture-recapture/resighting data. Each horizontal line connect capture or resightings of the same individual, a circle indicate that the individual is captured or resighted at least once during that year.
Model description:
The observations \(y_{it}\), an indicator of whether individual \(i\) was recaptured or resighted during year \(t\) is modeled conditional on the latent true state of the individual birds \(z_{it}\) (0 = dead or permanently emigrated, 1 = alive and at the study site) as a Bernoulli variable. The probability \(P(y_{it} = 1)\) is the product of the probability that an alive individual is recaptured or resighted, \(p_{it}\), and the state of the bird \(z_{it}\) (alive = 1, dead = 0). Thus, a dead bird cannot be recaptured or resighted, whereas for a bird alive during year \(t\), the recapture/resighting probability equals \(p_{it}\): \[y_{it} \sim Bernoulli(z_{it}p_{it})\] The latent state variable \(z_{it}\) is a Markovian variable with the state at year \(t\) being dependent on the state at year \(t-1\) and the apparent survival probability \(\phi_{it}\): \[z_{it} \sim Bernoulli(z_{it-1}\phi_{it})\]
We use the notation \(\phi\) to indicate that this parameter can be interpreted as apparent survival rather than true survival of which the standard notation would be \(S\) (D. L. Thomson et al. 2009). We model \(\phi\) dependent on age (1=first year, 2=older) and year.
\[\phi_{it} = \alpha_{age-class[it], t} \]
Annual recapture/resighting probability is modeled for each year independently: \[p_{it} = \beta_{t}\] We use uniform prior distributions for all parameters with a parameter space limited to values between 0 and 1 (probabilities), \(\alpha_{age-class,t} \sim uniform(0,1)\) and \(\beta_t \sim uniform(0,1)\).
We use Markov chain Monte Carlo simulations as implemented in jags (Plummer 2003) to fit the model to the data. For doing so, we first need to specify the model using Jags language. We do that in a separate text-file (jags/cjs.txt).
To fit the model, the Software jags need to be downloaded and installed.
## ## jags model code
## model{
## ## likelihood
## for(i in 1:nind){
## z[i,first[i]] <- 1
## for(t in (first[i]+1):nocc) {
## phiz[i, t-1] <- z[i,t-1]*phi[i, t-1]
## z[i,t] ~ dbern(phiz[i, t-1])
## zp[i,t] <- z[i,t]*p[i,t-1]
## y[i,t] ~ dbern(zp[i,t])
## }
## }
##
## ## linear predictors
## for(i in 1:nind){
## for(t in first[i]:(nocc-1)){
## phi[i,t] <- a[age[i,t],t]
## p[i,t] <- b[t]
## }
## }
##
## ## priors
## for(t in 1:(nocc-1)){
## b[t] ~ dunif(0, 1)
## a[1,t] ~ dunif(0, 1)
## a[2,t] ~ dunif(0, 1)
##
## }
## }After having specified the model, we need to bundle the data into a list. We have prepared that list in the object datax.
str(datax)
## List of 5
## $ y : 'table' num [1:1310, 1:9] 0 1 1 1 1 1 1 1 1 1 ...
## $ first: int [1:1310] 8 1 1 1 1 1 1 1 1 1 ...
## $ age : num [1:1310, 1:9] NA 2 2 1 1 1 1 2 1 1 ...
## $ nind : int 1310
## $ nocc : int 9We then need to specify initial values and the parameters that we would like to save.
library(R2jags)
initz <- matrix(NA, ncol=datax$nocc, nrow=datax$nind)
for(i in 1:datax$nind) initz[i,(datax$first[i]+1):datax$nocc] <- 1
initfun <- function(){
list(a=matrix(runif(2*(datax$nocc-1), 0,1), ncol=datax$nocc-1, nrow=2),
b=runif(datax$nocc-1, 0,1),
z=initz)
}
parameters <- c("a", "b")
mod <- jags(datax, inits=initfun, parameters.to.save=parameters,
model.file="jags/cjs.txt", n.chains=3, n.iter=2000)
mod <- mod$BUGSoutput
save(mod, file="modelfits/survivalmodel.rda")Before we look at the results, we check how well the Markov chains converged. The chains look ok except for the parameters of the last year. For the last year, apparent survival cannot be separated from recapture/resighting probability. Thus, we must expect that these chains do not converge well.
In the Cormack-Jolly-Seber model, for the last occasion, only the product of survival and recapture probability is estimable. Therefore, for these two parameters, it does not look like the Markov chains have converged.
We then look at the results
load("modelfits/survivalmodel.rda")
mod$summary
## mean sd 2.5% 25% 50%
## a[1,1] 3.568470e-01 0.14788623 1.405587e-01 2.539947e-01 3.321348e-01
## a[2,1] 3.755367e-01 0.19356107 8.713047e-02 2.282797e-01 3.459405e-01
## a[1,2] 3.865373e-01 0.13942289 1.550767e-01 2.900549e-01 3.737544e-01
## a[2,2] 5.488762e-01 0.13070284 3.242579e-01 4.544082e-01 5.434537e-01
## a[1,3] 5.486558e-02 0.05590406 1.886321e-03 1.593380e-02 3.655845e-02
## a[2,3] 8.190587e-01 0.11779720 5.571751e-01 7.413001e-01 8.370315e-01
## a[1,4] 1.729962e-01 0.10769660 2.468798e-02 9.276290e-02 1.545091e-01
## a[2,4] 5.173047e-01 0.05528124 4.118721e-01 4.789383e-01 5.164283e-01
## a[1,5] 4.055602e-01 0.14212213 1.609208e-01 3.004083e-01 3.955103e-01
## a[2,5] 7.329404e-01 0.06171004 6.143780e-01 6.904521e-01 7.336408e-01
## a[1,6] 1.363609e-01 0.06406940 3.885308e-02 8.852954e-02 1.254485e-01
## a[2,6] 5.955792e-01 0.03278355 5.299185e-01 5.742478e-01 5.952925e-01
## a[1,7] 8.791460e-02 0.08449543 2.056102e-03 2.461361e-02 6.273840e-02
## a[2,7] 4.525413e-01 0.02632604 4.028667e-01 4.333533e-01 4.520473e-01
## a[1,8] 4.298138e-02 0.04050236 1.271957e-03 1.224750e-02 3.034956e-02
## a[2,8] 7.443479e-01 0.12248904 4.961986e-01 6.554350e-01 7.602906e-01
## b[1] 3.970041e-01 0.15924497 1.363171e-01 2.770442e-01 3.758527e-01
## b[2] 4.108497e-01 0.10578283 2.134573e-01 3.371822e-01 4.063393e-01
## b[3] 5.674775e-01 0.10727348 3.541829e-01 4.938086e-01 5.699489e-01
## b[4] 7.183032e-01 0.06594685 5.854001e-01 6.725983e-01 7.206398e-01
## b[5] 6.373483e-01 0.05824699 5.216230e-01 5.983343e-01 6.393034e-01
## b[6] 8.787225e-01 0.03299766 8.101230e-01 8.562140e-01 8.808235e-01
## b[7] 7.285209e-01 0.03813321 6.511608e-01 7.025155e-01 7.297285e-01
## b[8] 5.984491e-01 0.11120362 4.534393e-01 5.161797e-01 5.666464e-01
## deviance 1.332716e+03 135.67941260 1.014992e+03 1.256716e+03 1.362579e+03
## 75% 97.5% Rhat n.eff
## a[1,1] 4.331101e-01 0.7264191 1.015359 290
## a[2,1] 4.955586e-01 0.8197887 1.004035 2000
## a[1,2] 4.692237e-01 0.7033673 1.001147 3000
## a[2,2] 6.369562e-01 0.8146704 1.016822 130
## a[1,3] 7.765612e-02 0.2033934 1.002384 3000
## a[2,3] 9.111923e-01 0.9903918 1.004871 460
## a[1,4] 2.316744e-01 0.4308784 1.001124 3000
## a[2,4] 5.546029e-01 0.6297880 1.007154 380
## a[1,5] 5.069964e-01 0.6992618 1.004664 480
## a[2,5] 7.737150e-01 0.8562529 1.012477 170
## a[1,6] 1.739418e-01 0.2904621 1.001019 3000
## a[2,6] 6.169479e-01 0.6609227 1.010869 210
## a[1,7] 1.235419e-01 0.3074530 1.002424 1000
## a[2,7] 4.707361e-01 0.5044802 1.006162 360
## a[1,8] 6.149474e-02 0.1465196 1.003023 870
## a[2,8] 8.398049e-01 0.9450982 1.403860 9
## b[1] 5.019517e-01 0.7367122 1.008187 320
## b[2] 4.836419e-01 0.6240565 1.005672 390
## b[3] 6.450357e-01 0.7738967 1.004918 450
## b[4] 7.659016e-01 0.8364106 1.006271 670
## b[5] 6.775658e-01 0.7491825 1.001338 2400
## b[6] 9.027881e-01 0.9366115 1.014192 160
## b[7] 7.544946e-01 0.7980943 1.004610 600
## b[8] 6.604551e-01 0.8672611 1.399615 9
## deviance 1.427505e+03 1531.6642401 1.412915 9plot(c(2015,2023), c(0,1), type="n", las=1, xlab=NA, ylab="Apparent survival")
phi <- apply(mod$sims.list$a, c(2,3), mean)
phi.lwr <- apply(mod$sims.list$a, c(2,3), quantile, probs=0.025)
phi.upr <- apply(mod$sims.list$a, c(2,3), quantile, probs=0.975)
x <- seq(2015.5, 2022.5, by=1)
segments(x, phi.lwr[1,], x,phi.upr[1,])
points(x, phi[1,], pch=21, bg="white")
segments(x+0.1, phi.lwr[2,], x+0.1,phi.upr[2,], lwd=2, col="brown")
points(x+0.1, phi[2,], pch=21, bg="white", col="brown")
legend(2015, 1.1, xpd=NA, lwd=c(1,2), col=c(1,"brown"), legend=c("first year", "adult"), horiz=TRUE)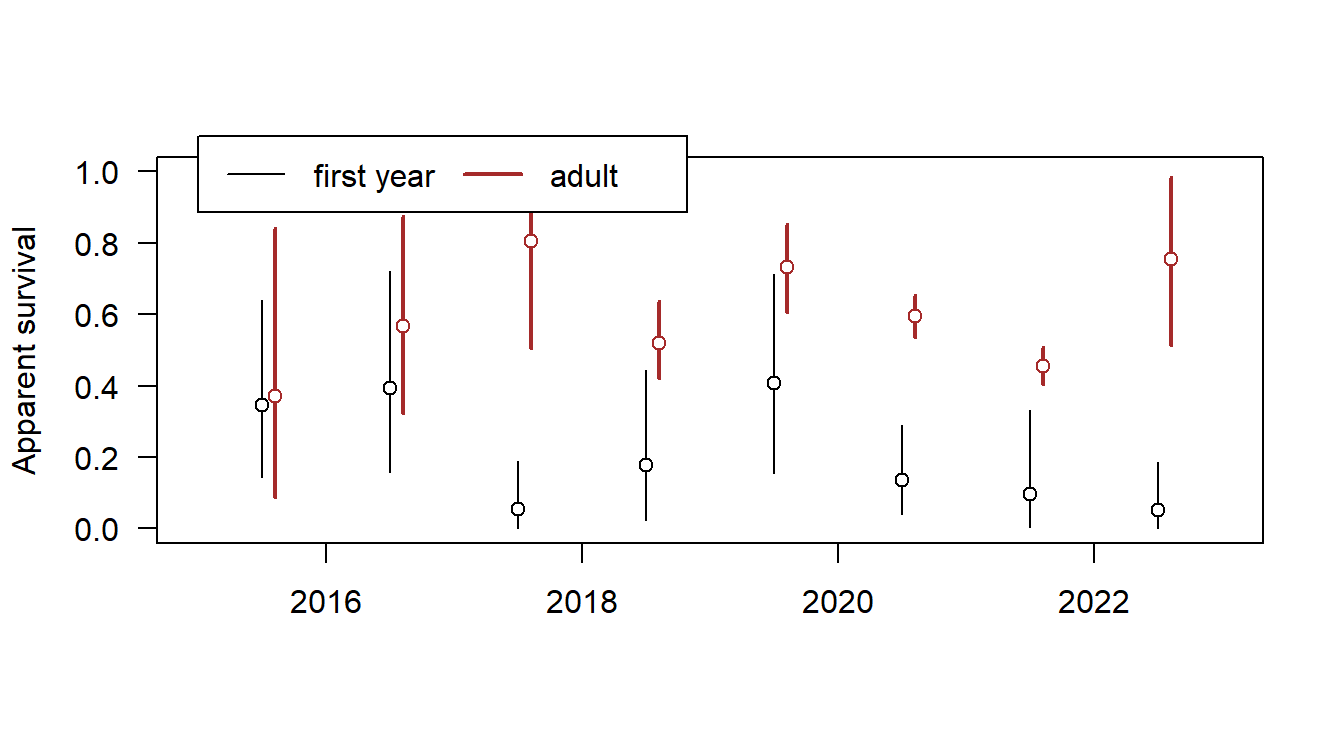
Figure 17.2: Annual apparent survival of juvenile and adult Snowfinches.
plot(c(2016,2023), c(0,1), type="n", las=1, xlab=NA, ylab="Recapture/resighting probability")
p <- apply(mod$sims.list$b, 2, mean)
p.lwr <- apply(mod$sims.list$b, 2, quantile, probs=0.025)
p.upr <- apply(mod$sims.list$b, 2, quantile, probs=0.975)
x <- seq(2016, 2023, by=1)
segments(x, p.lwr, x,p.upr, lwd=2, col="brown")
points(x, p, pch=21, bg="white", col="brown")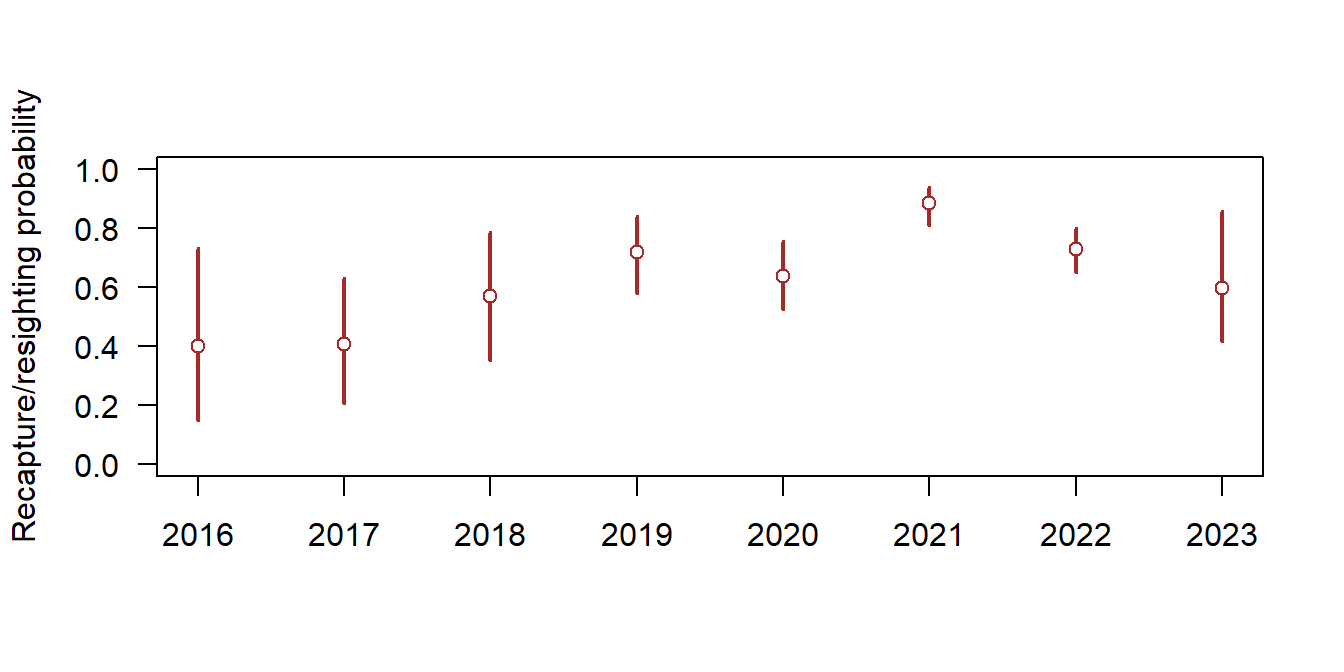
Figure 17.3: Recapture and resighting probability
Kéry and Schaub (2012) wrote a gentle introduction to mark-recapture modelling for biologists. An example using Stan is found here https://tobiasroth.github.io/BDAEcology/cjs_with_mix.html.